Masks and Region Of Interests (ROIs)
Author: Syeda Warda Taqdees
Contributor/s: Dr. Christina Tuke Flanders
Learning Objectives
After completing this lesson, students should be able to
- Select an elliptical ROI manually and display it.
- Manually select a polygonal ROI and adjust its shape interactively.
- Create draggable freehand region on MRI slice data.
- Sequentially delineate ROIs in multiple MRI slices.
- Select and store multiple ROIs on a single image.
- Learn to use 'regionprops' for measuring properties of image regions.
- Extract brain and perform skull removal on brain MRI data semi-automatically.
Manual Image Masks
MATLAB's image processing toolbox provides a variety of tool for manually selecting an image ROI.
Elliptical ROIs
'imellipse()' allows to manually place an ellipse on the image and returns the handle to the 'ellipse' object. The method createMask accepts the ellipse object handle and creates a mask within the image. A mask is a binary image (logical) with 1's assigned to the voxels within ROI and 0's assigned to the voxel outside ROI.
% Manual ROI selection
% Displaying Figure
figure;
imagesc(mriImage1);
% Manually creating elliptical ROI binary mask
hElips = imellipse();
elipsBW = createMask(hElips);
Next we display both Mask image and the selected ROI. We can extract ROI voxels from the image by carrying out voxel-wise multiplication with the binary mask. Notice that we convert 'logical' mask to 'double' to ensure compatibility between mask and image datatypes.
% Displaying binary mask
figure
subplot(1,2,1)
imagesc(elipsBW); colormap('gray')
axis image
% Displaying selected ROI
subplot(1,2,2)
imagesc(double(elipsBW).*mriImage1)
axis image
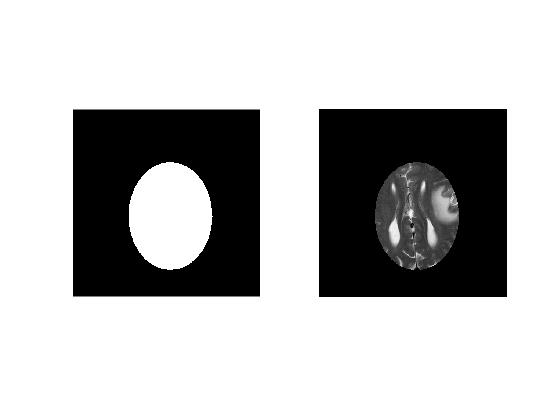
Polygonal ROIs
'roipoly()' allows us to interactively select a polygonal ROI on image. Every time a click is made on the image, a vertex of the polygon is created at that position. 'roipoly' returns a logical binary mask
% Manual ROI selection
% Displaying slice
figure
imagesc(mriImage1);
% Manually selecting ROI using roipoly
polyBW = roipoly();
% Display selected ROI Mask
figure
subplot(1,2,1)
imagesc(polyBW); colormap('gray')
axis image
% Disply selected ROI
subplot(1,2,2)
imagesc(double(polyBW).*mriImage1); colormap('gray');
axis image
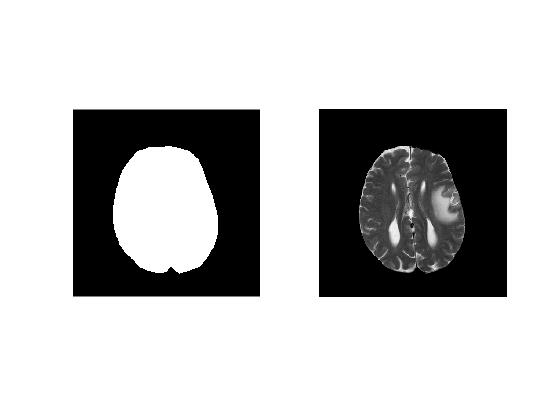
So far, we have been creating masks on a single slice. Recall that our MRI image dataset consists of multiple slices. All manual selection techniques explained previously can be used in a loop to create binary masks for each slice in the dataset. Note that it is important to save slice mask in each iteration separately. Using this trick, we create a 3D volume mask for our data.
% roipoly can be used in a loop to create masks for multiple slices
figure
iMask = 1;
for iImg = [1:10:22]
% Selecting and displaying slice
mriSlice = mriImage(:,:,iImg);
imagesc(mriSlice); colormap('gray');
% Creating binary mask
polyBW(:,:,1,iMask) = roipoly();
% Selecting roi
sliceROI(:,:,1,iMask) = double(polyBW(:,:,1,iMask)).*mriImage(:,:,iImg);
% incrementing mask index
iMask = iMask+1;
end
% Display mask image
figure;
montage(polyBW);
% Display Selected ROIs
figure;
montage(sliceROI,gray(1000));
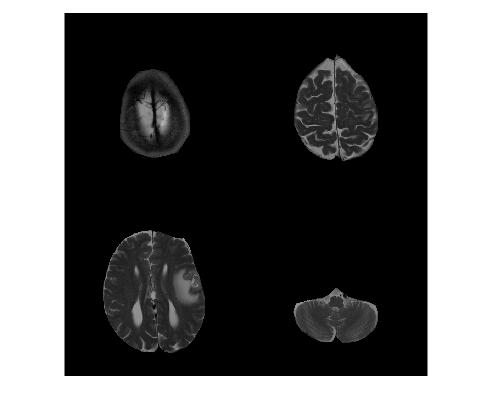
Challenge!
% Challenge 1
% Using the downloaded dataset of your choice, manually delineate ROIs in
% first 5 slices.
% Display montage of masks created in previous step.
% Display montage of selected ROIs.
% Extension: By building upon the concept of looping, create binary masks
% of multiple ROIs in same slice e.g. masks for eyes, brain etcFreehand ROIs
'imfreehand' is a handy function to draw draggable freehand ROIs on an image. It works similar to 'imellipse' and returns a 'freehand ROI' object which can later be used to create a mask by calling 'createMask' method.
% imfreehand can also be used to manually select an ROI
% Displaying slice for ROI selection
figure;
imagesc(mriImage1);
% selecting and creating ROI mask
hFree = imfreehand();
freeBW = createMask(hFree);
% Displaying binary mask
figure
subplot(1,2,1)
imagesc(freeBW); colormap('gray')
% Displaying selected ROI
subplot(1,2,2)
imagesc(double(freeBW).*mriImage1);colormap('gray');
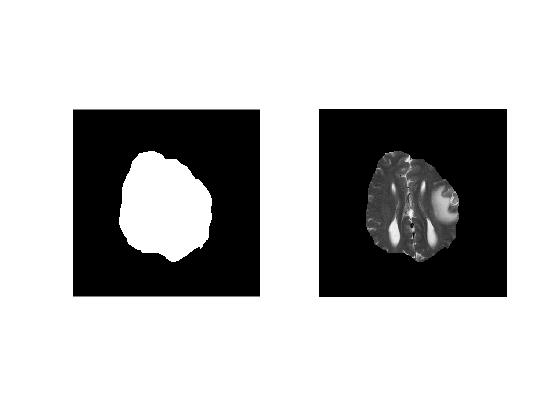 Challenge!
Challenge!
% Challenge 2
% By using the concepts of 'for loop', select 2-3 ROIs in a single slice.
% Hint: delete(h) deletes the perviously drawn region
% Merge all binary masks into a single mask. Hint: Check out 'sum' function
% Extension: Colorcode each selected ROI by assigning a different number to
% voxels in each binary mask.Semi-automatic ROI Selection
MATLAB image processing toolbox provides useful fucntions for automating ROI selection in MRI images. In this section, we are going to utilize these functions for brain extraction.
Automatic Ellipse Placement
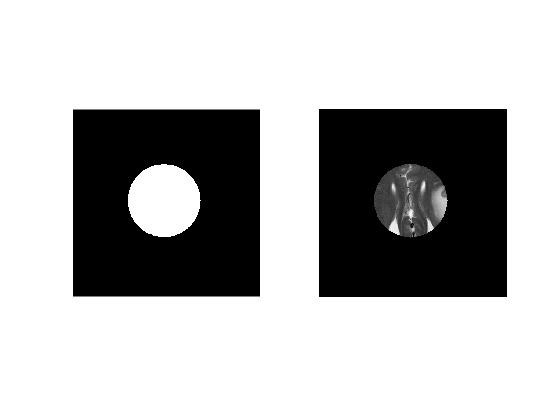
'imellipse()' also accpets position and size of an elliptical object that can be automatically placed on an image.
% semi-automatic ROI selection
% Automatic ROI selection using imellipse
figure;
imagesc(mriImage1);
% Creating elliptical binary mask
elipsAutoPosition = [150 150 200 200];
autoElipseBW = createMask(imellipse(gca,elipsAutoPosition));
% Displaying binary mask
figure
subplot(1,2,1)
imagesc(autoElipseBW); colormap('gray')
% Displaying selected ROI
subplot(1,2,2)
imagesc(double(autoElipseBW).*mriImage1);colormap('gray');
Semi-automatic Brain Extraction
In brain extraction pipeline, first step is to normalize image slice voxels.
Normalising MRI Image
% Semi-automatic Brain Extraction
% Normalizing MRI image
normMriImage1 = mriImage1./max(mriImage1(:));
Selecting Binarization Threshold
In order to create a binary mask for brain region, we need to select an optimal binarization threshold. It means that if the value in a voxel is greater than the threshold values then it will be considered as a brain voxel, otherwise it will be considered an outside voxel.
For selecting an optimal threshold, we create histogram of normalized image intensities.
% histogram of normalized MRI image
figure;
imhist(normMriImage1);
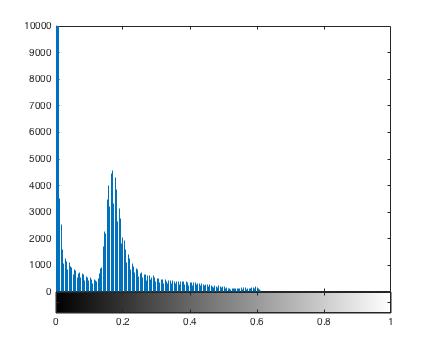 Notice that the low intensity background voxels are roughly grouped in the intensity range 0-0.15.
Notice that the low intensity background voxels are roughly grouped in the intensity range 0-0.15.
To check spatial distribution of intensities, I find it useful to create a slice mesh plot.
% Displaying Mesh plot of slice
figure;
mesh(normMriImage1);
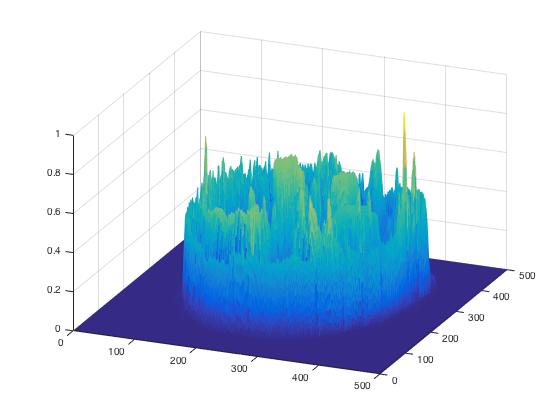
Based on historgram and meshplot, we select a threshold of 0.15 and create a binary mask.
% Creating binary mask
normBW = im2bw(normMriImage1,0.15);
% Displaying binary mask
figure
subplot(1,2,1)
imagesc(normBW); colormap('gray')
% Displaying selected ROI
subplot(1,2,2)
imagesc(double(normBW).*normMriImage1);colormap('gray');
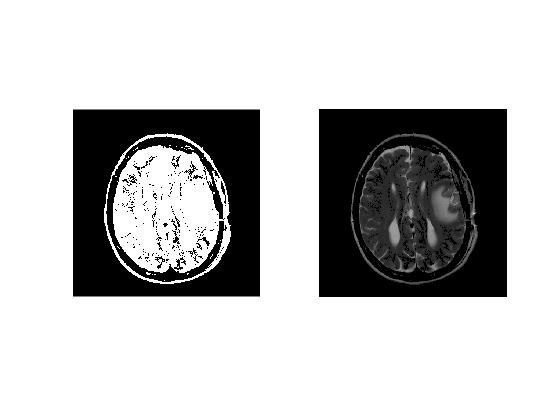
Cleaning Binary Mask
We use 'bwareaopen' to remover all isolated regions of size 500 or less.
% clean mask of small areas
cleanBW = bwareaopen(normBW,500);
% Displaying binary mask
figure
subplot(1,2,1)
imagesc(cleanBW); colormap('gray')
% Displaying selected ROI
subplot(1,2,2)
imagesc(double(cleanBW).*normMriImage1);colormap('gray');
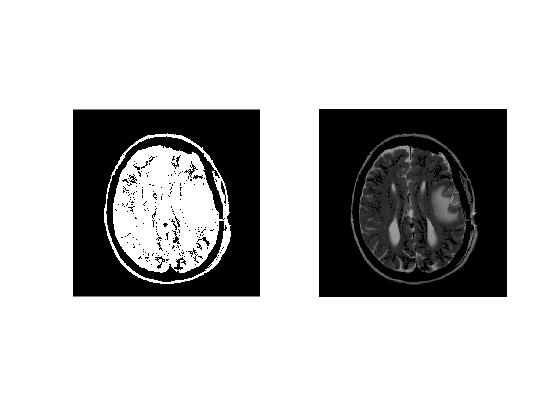
Selecting Brain Region
To select the brain regions, we calculate the area of each isolated region by using 'regionprops' and select the region with largest area as brain region.
To do this, we assign a unique label to each region by using 'bwlabel'
% Label Regions
labelBW = bwlabel(cleanBW);Next, we calculate area of each labeled region and find the index of the region with largest area.
% Calculate number of voxels in each region
regStats = regionprops(cleanBW,'area');
allAreas = [regStats.Area];
% Find region with largest area
[brainArea brainInd] = max(allAreas);
Finally, we extract the largest region by using 'ismember'.
% Extract the largest region using ismember()
brainRegion = ismember(labelBW, brainInd);
% Convert from integer labeled image into binary image.
brainBW = brainRegion > 0;To fill in any holes left in the region due to binarization, we use 'imfill' function.
% Fill holes in brain mask
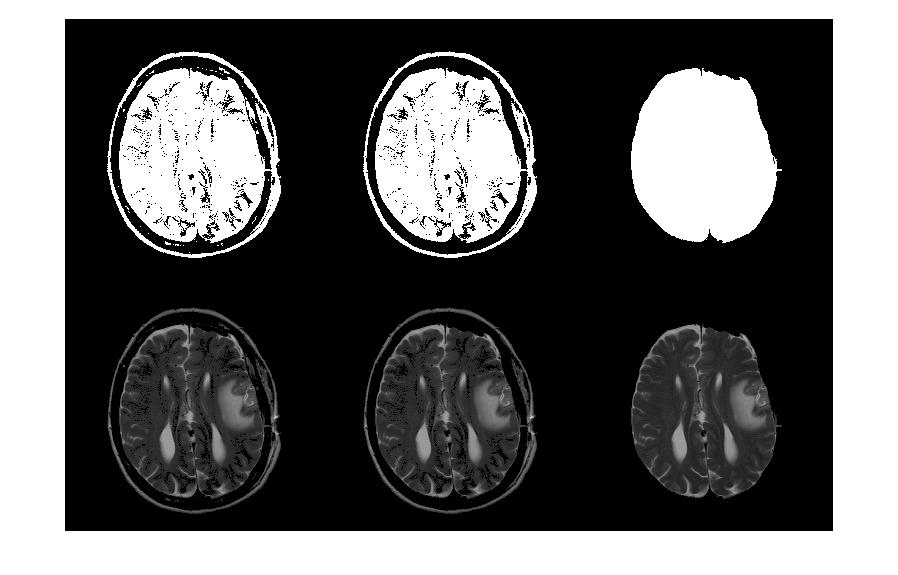
fillBrainBW = imfill(brainBW,'holes');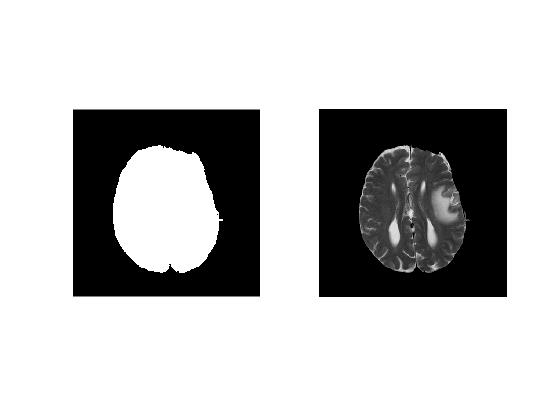 A summary of semi-automatic brain extraction pipeline is presented in the montage below.
A summary of semi-automatic brain extraction pipeline is presented in the montage below.
 Challenge!
Challenge!
% Challenge 3
% Select slice# 15 of this T2 brain dataset and create a normalized image
% Perform brain extraction by adjusting parameters